Sunday, 29 Jan 2023
Automating bioinformatic data to help end world hunger

In kommit, we succeeded in supporting the automation and data management strategy for a big-impact genetic research center. By optimizing complex bioinformatic processes, we saved a team of renowned scientists countless hours of work. Their goal? Making crops more resistant to weather and climate change to help end the food crisis — and ours, help them do it more efficiently.
“One machine can do the work of fifty ordinary men. No machine can do the work of one extraordinary man,” said Elbert Hubbard more than a century ago.
Although no artificial intelligence or software data automation existed during the life of this known American writer, he had a point.
At kommit, we’ve never pretended to be experts in the complexity of our client's industries or products, less in bioinformatics (or bioinformatic pipelines for that matter) — nor have we wanted to replace in any way the bioinformatician professionals in that field that reached us for help about six months ago.
But we understood how data management automation could take their advanced genetic research project to a new level, a bioinformatic endeavor called Elastic Search Implementation for Rising Genome Analytics (ESIRGA), with the following outcomes:
- Faster data interpretation and visualization
- Improved data reliability and utility
Certainly, software automation is everywhere. According to Bots & People, “the global Robotic Process Automation market is projected to hit 23,9 Million US Dollars in 2030”. If there’s any time-consuming process in a business or market that’s worth automating, never doubt it will be.
So we had a unique opportunity to leverage our developers’ expertise in this field by owning the optimization of the bioinformatic analysis area of ESIRGA. Our main goal was to provide the underlying genetic data processes with more efficient storage, processing, and retrieval of data, files, and information.
How did we face such a challenge from the software engineering and automation perspective, and made scientists’ lives easier in their quest to make plants more resistant to weather changes and crops endure different types of conditions?
Find out about it below, in the first entry of kommit’s new blog. We hope you enjoy it.
Table of contents
Data Management and Bioinformatics
kommit’s experience with ELK and data
kommit and bioinformatic projects
Data Management and Bioinformatics
The good thing about bioinformaticians is that they know what they’re up to. Their expertise relies on managing specialized raw reads to extract complex information for scientific analysis.

However, since they are not software experts, many processes in what they call bioinformatic pipelines are done manually, consuming the time needed for other tasks like interpreting results, gaining insights into diseases, and coming up with treatment plans.
That’s where kommit stepped in to help, putting together a software engineering team of five full-time dedicated developers to build a customized solution to automate the scientist’s procedures in these pipelines.
The road to automation
First, our six-month partnership with the client began by figuring out which type of data the research team extracted during the bioinformatic pipeline, starting from the Genome sequencing and assembly — the complete analysis of DNA sets of a genome and their translation to FASTA files.
This leads scientists to what is known as genome annotation, a three-part process of manual tabulation of different DNA sequences and their corresponding files:
Transposable element (TE) annotation to identify and classify DNA sequences with complex patterns, under the *.te file format.
Gene Annotation to identify and classify the genes inside the genome, resulting in .gff files.
Variant calling, which is the process of identification of mutations in the genome. The final files in this step come out in the .vcf format.

As you can see, these activities create several types of files, each one with a different management system. This, in turn, challenges the researcher when trying to, first, interpret results, and then present them with data visualization.
We tackled such a complex and time-consuming process by automating three main activities:
- “The genome assembly and quality analysis”
- “The DNA data search in large files”
- “The visual DNA comparison among genes of several individuals of the same species”
How did we make it work?
For genome assembly and quality analysis, kommits’ team implemented a suite of orchestration scripts on Ruby that enabled bioinformaticians to drag and drop raw files into a folder where the corresponding pipeline executes automatically.

Afterward, we built an application in Ruby on Rails to extend script functionalities and ease the pipeline execution “In the UI, the user enters a path to the file which will be processed, and the application copies it to another folder where a Linux cronjob searches for new files and starts the pipeline execution.”
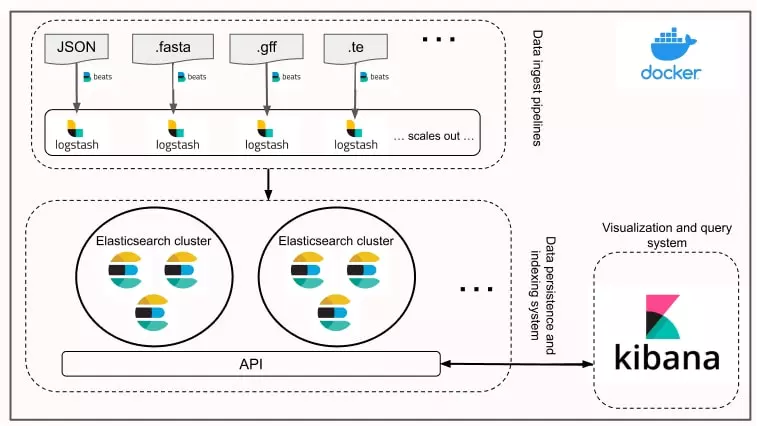
We also implemented a custom-built indexing system using the Elasticsearch, Logstash, Beats and Kibana (ELK) stack to ease the queries in large files using non-relational databases.
Delivering tangible results
The ESIRGA project is currently in its early stages, and the research team is still experimenting with automation. One of our most important developments was reducing the workload for the bioinformatician by 20%.
Now, they can just search for a gene or mutation by typing its name or some features, and customized visualizations of their data just pop up.
Also, after our implementation, the scientific project showcased the tool at the 19th International Symposium on Rice Functional Genomics (ISRFG 2022), as “Rising Genome Analytics to Elastic Search Implementation for Rice Genome Analytics”.
ELK expertise at kommit
The ELK stack is one of the most used stacks for real-time data extraction, recompilation, and visualization for enterprise solutions.
As a software engineering company that delivers tech and innovation, we already had experience in data management with ELK, which prove us to be very powerful in solving the ESIRGA project’s challenges thanks to the following features:
- High performance
- Easily scalable
- Distributed architecture
- Document-oriented database
- Schema free
- API-driven
- Real-time search engine
- Multi-tenancy
The next diagram illustrates how we used the ELK stack to index, access, and retrieve data from the biological research performed by the ESIRGA team.
Our future with bioinformatic projects
Now that you know about how we automated the bioinformatic processes of ESIRGA, one of the world’s main projects tackling world hunger through genetic engineering, we’re glad to share that we’ll continue moving in this direction in the future.
We’re already working on ESIRGA’s second phase, which entails the implementation of a module that will integrate the system with GenBank, and another module that integrates with the high-performance machines that power the project.
Keep in touch if you have any questions or wish to know about kommit’s commitment to enterprises with the potential of changing the world as we know it!
And to say goodbye (for now), a famous phrase by Tom Preston-Werner: “You’re either the one that creates the automation or you’re getting automated.”
Written by: kommit